Extensive personalized human gut strain collection
with phenotypic data by CAMII
Contains 16584 isolates spanning >400 taxa with a rich set of linked
morphologic, phenotypic, taxonomic and WGS data
Culturomics by Automated Microbiome
Imaging and Isolation (CAMII) system
CAMII enables large-scale end-to-end generation of clonal
strains from diverse input microbiome samples.
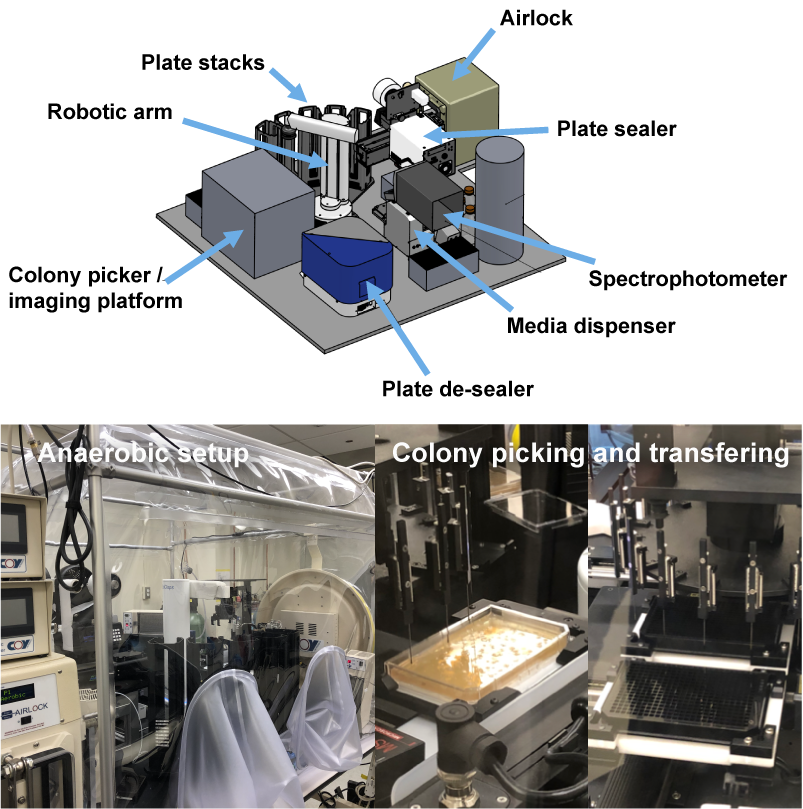
Maximum capacity: 2,000 colonies per hour and 12,000 isolates per run
Standardized colony morphological data
gathering and analyzing
Morphological features of colonies significantly improved
strain isolation and cultivation
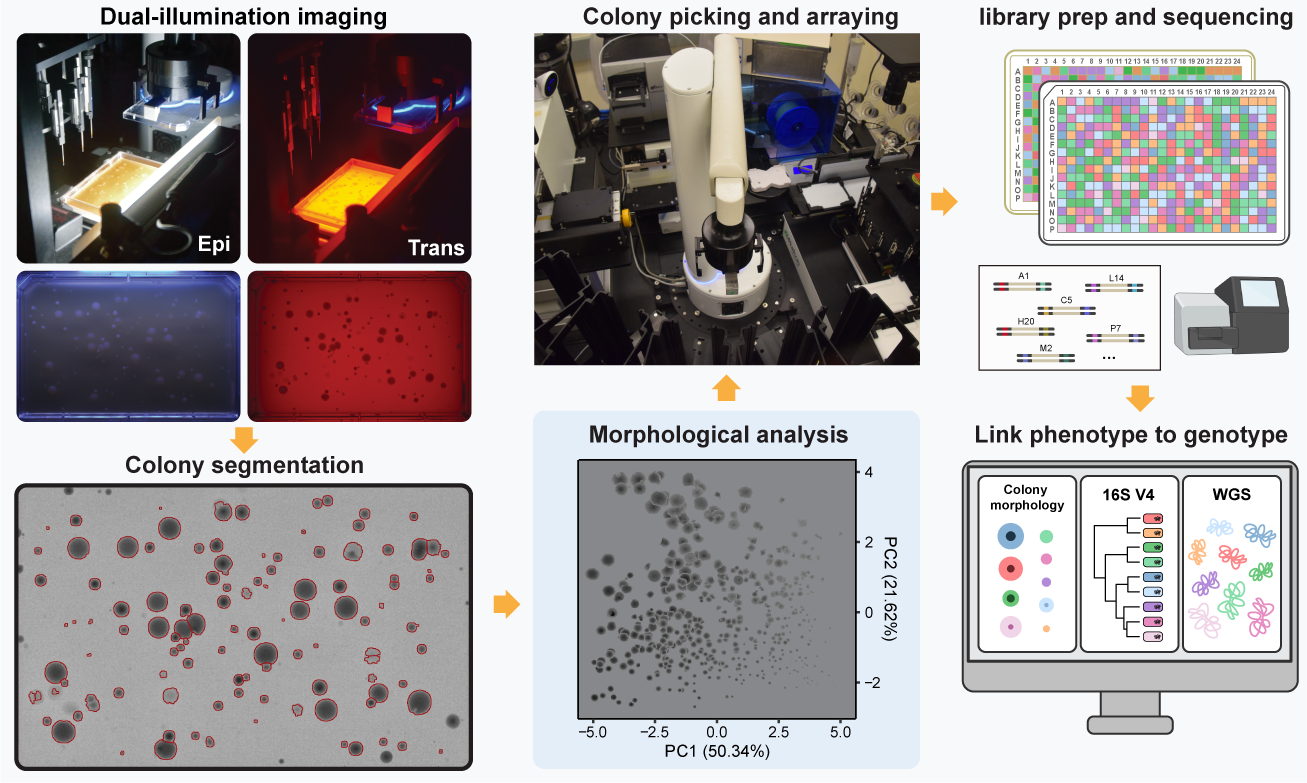
Scripts for analyzing colony images can be accessed at GitHub

Summary of biobanks
Summarize isolates information across different individuals in CAMII biobanks.
Search by Taxonomy
Search isolates by specific taxa or best match to provided 16S-V4 sequence
Personalized gut isolates generated by CAMII
About isolates request
There are several ways to request specific isolates or collections generated by CAMII
Search by Isolate
Check the information of all isolates by keywords
Information of all isolates
| Isolate ID | Destination plate | Destination well | Culturing media | Source plate | Source index | Source sample | Isolation platform | Isolation strategy | Isolation date | Taxonomy by 16SV4 | Whole genome sequencing | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Kindom | Phylum | Class | Order | Family | Genus | Species | Purity (%) | Hetergenity (%) | Assembly size (bp) | # of contigs | Largest contig (bp) | N50 (bp) | GC content (%) | # of PE reads | 16SV4 sequence | |||||||||
| Isolate ID | Destination plate | Destination well | Culturing media | Source plate | Source index | Source sample | Isolation platform | Isolation strategy | Isolation date | Taxonomy by 16SV4 | Whole genome sequencing | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Kindom | Phylum | Class | Order | Family | Genus | Species | Purity (%) | Hetergenity (%) | Assembly size (bp) | # of contigs | Largest contig (bp) | N50 (bp) | GC content (%) | # of PE reads | 16SV4 sequence | |||||||||
Search by Taxonomy
Search isolates by specific taxa or find isolates that best match 16S sequence your provide
| Isolate ID | Destination plate | Destination well | Culturing media | Source plate | Source index | Source sample | Isolation platform | Isolation strategy | Isolation date | Taxonomy by 16SV4 | Whole genome sequencing | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Kindom | Phylum | Class | Order | Family | Genus | Species | Purity (%) | Hetergenity (%) | Assembly size (bp) | # of contigs | Largest contig (bp) | N50 (bp) | GC content (%) | # of PE reads | 16SV4 sequence | |||||||||
| Matched sequence | Percent identity | Isolate ID | Destination plate | Destination well | Culturing media | Source plate | Source index | Source sample | Isolation platform | Isolation strategy | Isolation date | Taxonomy by 16SV4 | Whole genome sequencing | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Kindom | Phylum | Class | Order | Family | Genus | Species | Purity (%) | Hetergenity (%) | Assembly size (bp) | # of contigs | Largest contig (bp) | N50 (bp) | GC content (%) | # of PE reads | 16SV4 sequence | |||||||||||
Search by Morphology
Search isolates by specific morphological features
| Isolate ID | Destination plate | Destination well | Culturing media | Source plate | Source index | Source sample | Isolation platform | Isolation strategy | Isolation date | Taxonomy by 16SV4 | Whole genome sequencing | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Kindom | Phylum | Class | Order | Family | Genus | Species | Purity (%) | Hetergenity (%) | Assembly size (bp) | # of contigs | Largest contig (bp) | N50 (bp) | GC content (%) | # of PE reads | 16SV4 sequence | |||||||||
Download CAMII datasets
16S taxonomy, WGS and morphological features
Datasets of all isolates:
Datasets of all colonies visually captured: